I thought it would be fun to try analyzing the mtDNA sequence of the presumed Richard III skeleton using my mthap program to see what it comes up with.
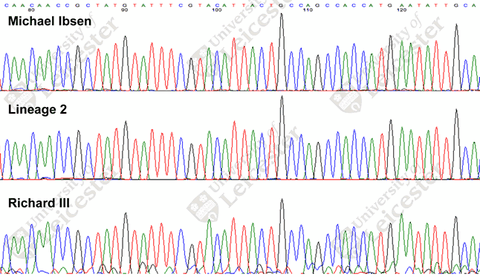
Unfortunately, only a 53bp fragment of the ~16569bp mtDNA sequence has been released so far, via this image on the University of Leicester web site:
This has been transcribed thanks to Ann Turner and Dave Cissell on the ISOGG list as:
CAACAACCGCTATGTATTTCGTACATTACTGCCAGCCACCATGAATATTGCA
This covers the sequence from positions 16076 to 16127 relative to the rCRS or RSRS reference sequences. (The position numbers on the image are presumably offset due to an earlier insertion.) The only difference to rCRS in the sequence is 16126C. That’s not a lot to work with. Even a basic HVR1 mtDNA test covers over 500 positions sequenced and can still be highly ambiguous. These days a solid haplogroup assignment requires a full sequence, or at least a genotype test covering thousands of markers across the full sequence to get you close (e.g. Genographic 2.0 or 23andMe).
But let’s see how far this gets us anyways.
Published reports indicate that Michael Ibsen, a descendant of Richard III’s mother is in haplogroup J but doesn’t get more specific. 16126C is one of the defining markers for the parent group JT, but it is also found in 11 other haplogroups in the current PhyloTree Build 15 reference. One of the defining markers for J is 16069T, but regrettably the image cuts off just a few bp shy of that marker, so at best this fragment can show JT, but not J.
We’d next want to go through the other 11 possible haplogroups to see which ones fit our fragment and which don’t. In other words, we’d want to look for which of these haplogroups have 16126C but not any other marker in the range of 16076 to 16127. We could do this manually, but that’s what computers are for, so let’s try to do this in an automated way.
I have a web application I’ve written called mthap which is freely available to use for analyzing mtDNA results. The basic version is very easy to use. Just feed it your data file and look at the results. That makes a lot of assumptions though, assumptions which don’t work well with a short fragment like this. Those assumptions are based on the most common DTC mtDNA tests. There’s also the less well known advanced options page where you can twiddle various parameters for unusual cases like this.
Here’s what we need to do:
- Make a text file with a text editor such as notepad with this content: 16126C
- Select this file on the “File:” line.
- Tick the HVR1 option.
- Enter 16076 to 16127 as the HVR1 positions.
- Click “Upload”
After waiting 10-15 seconds you will see the results. Due to the limitations of this sample, we get a lot of potential matches, but the first four are the most significant: H14b1, HV0a1, R0a, and finally JT. The rest are all either poor matches or subgroups of these. So we’ve managed to reduce our possibilities from 11 to 4, which is not bad. But we’re still left with a good deal of ambiguity. Presumably we will get a full sequence or at least a longer sequence once the scientific journals publish the full articles on the analysis.
UPDATE: See also the followup article: Analyzing the mtDNA of descendants of Richard III’s mother.
(Please note that this is not meant to cast doubt on the identification. It is merely to show how much objective data we can determine based on the currently released information.)
Due to increasing amounts of spam comments, comments for this post have been disabled.